FITMUNK
ACCELERATE REFINEMENT
Fitmunk provides a framework for fitting conformations on a fixed backbone into electron density.
Current Fitmunk applications can assist in protein model building, refinement and validation.
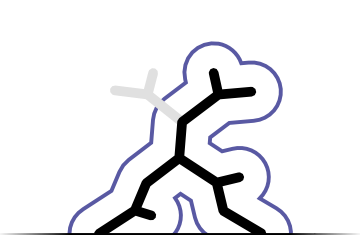
Conformation recogniton
Primary application of Fitmunk is a recognition of side chain conformations. Fitmunk can correctly recognize conformation at almost 100% of positions.
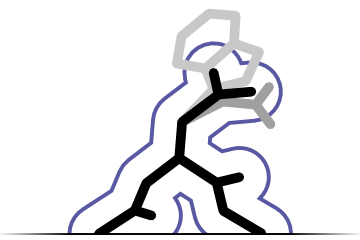
Sequence recognition
Fitmunk can recognize protein sequence from electron density maps without prior information about correct sequence. It can also assign a target sequence to a model. De novo sequence recognition is currently limited to topology recognition (e.g. no differentiation between ASN and ASP)

Alternative conformations
Fitmunk can model alternative conformations of amino acid sidechains. This feature is currently under development and the results may contain false positives
Porebski PJ, Cymborowski M, Pasenkiewicz-Gierula M, Minor W (2016)
Fitmunk: improving protein structures by accurate, automatic modeling of side-chain
conformations.
Acta Crystallogr D Struct Biol
72: 266-80.
[DOI:10.1107/S2059798315024730] [Pub Med ID: 26894674] [Pub Med Central ID: PMC4756610]